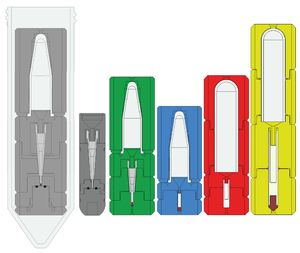
3D-Printable Centrifugal Rotor Packing Devices
|
3D-printable device assemblies for packing semi-solid samples into solid state NMR rotors using a centrifuge. Available for various rotor sizes and styles, including for 0.7 mm Bruker rotors. Intended to be printed using resin stereolithography (SLA) 3D-printing. Freely available for academic use, but please be sure to cite our article published in the Journal of Magnetic Resonance. If you have any questions about printing or modifying these designs, please reach out to Prof. Osborn Popp.
Files available on GitHub. |